I am a physician-scientist passionate about harnessing genomics to unravel the mechanisms driving human biology and disease. My career has evolved from hands-on bench work to the forefront of big data and computational analysis, with a continuous focus on chromatin organization. Ultimately, I strive to translate scientific discoveries into advances that deepen our understanding - and improve the diagnosis - of complex diseases.
During my graduate studies, I was honored to receive the NIH F31 fellowship under the mentorship of Drs. Rajan Jain and Jon Epstein. Together, we investigated how chromatin architecture controls gene regulation during cell differentiation, leading to several publications, including two first-author papers in journals like Nature Genetics and Circulation.
Now, as I pursue a career in pathology, I’m excited to apply my expertise in genetics, genomics, and computational biology to advance personalized medicine and improve patient care.
Outside of work, family is my priority. You’ll often find me chasing after my daughter at the neighborhood playground, exploring trails on family camping trips, or discovering new dining spots around Atlanta.
Download my curriculum vitae .
Access my genomics analysis Shiny server.
Explore Hi-C data in my HiGlass server .
- Genetics & Epigenetics
- Chromatin organization
- Computational genomics
- Data analysis & visualization
- Disease biology
MD, Pereleman School of Medicine, 2023
University of Pennsylvania
PhD, Genetics and Epigenetics, 2021
University of Pennsylvania
BS, Molecular and Cellular Biology, 2012
The Johns Hopkins University
Computational Skills
Shell Scripting | CLI | Sys Adm
Statistical Analysis | Visualization | R Shiny
Text Processing | Data Formating | Regex
Data Generation & Analysis | Bioconda
Research Experience
Featured Publications
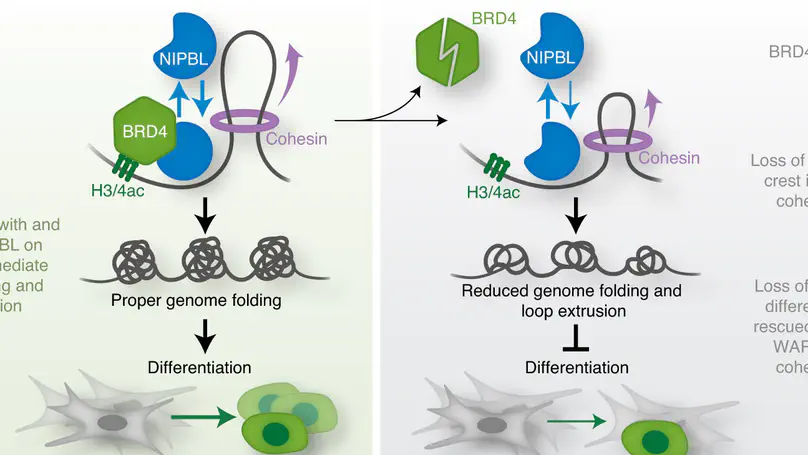
Here we define a role for bromodomain-containing protein 4 (BRD4) in 3D genome organization and its interaction with the cohesin loader NIPBL. Results of our experiments demonstrate that BRD4 depletion results in decreased NIPBL binding to cromatin, compromised genome folding and loop extrusion, and impared cell differentiation.
